-
Description
lncRNA (long non-coding RNA) is a kind of non-coding RNA with a length of more than 200 nt, which widely exists in organisms. TIANGEN uses a strand-specific method without rRNA for library construction. Through high-throughput sequencing technology, it obtains lncRNA sequence information with single base resolution. Relying on a powerful biological information analysis platform and a professional database, it identifies known lncRNA and searches for new lncRNA and target gene prediction.
-
Technical Workflow
Scheme design → Sample processing → Library construction sequencing → Data analysis → After-sales service
-
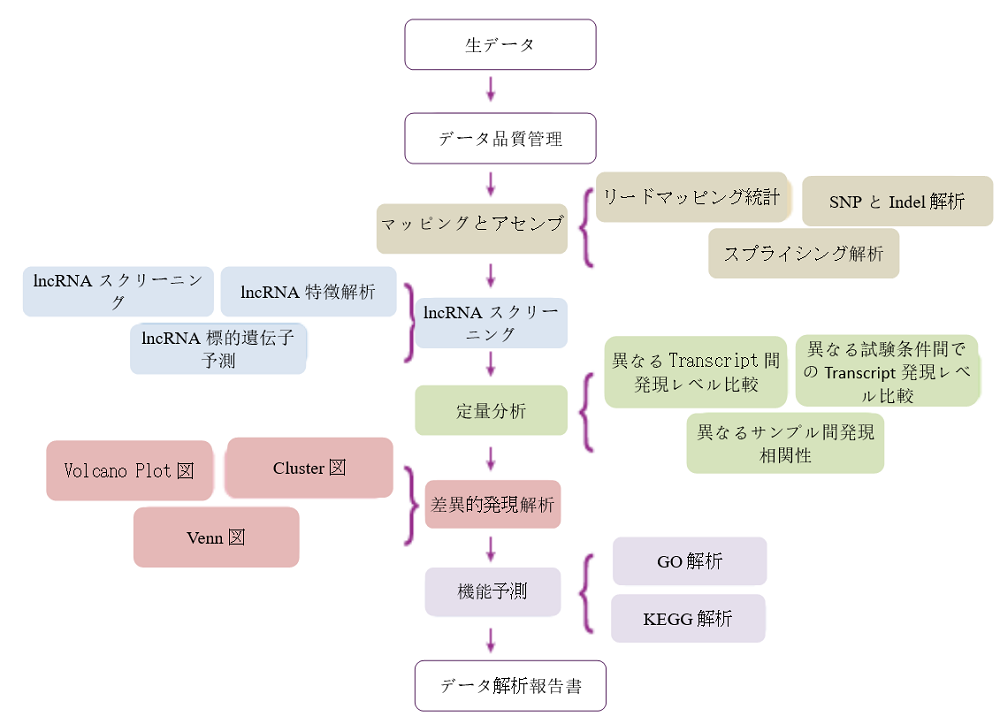
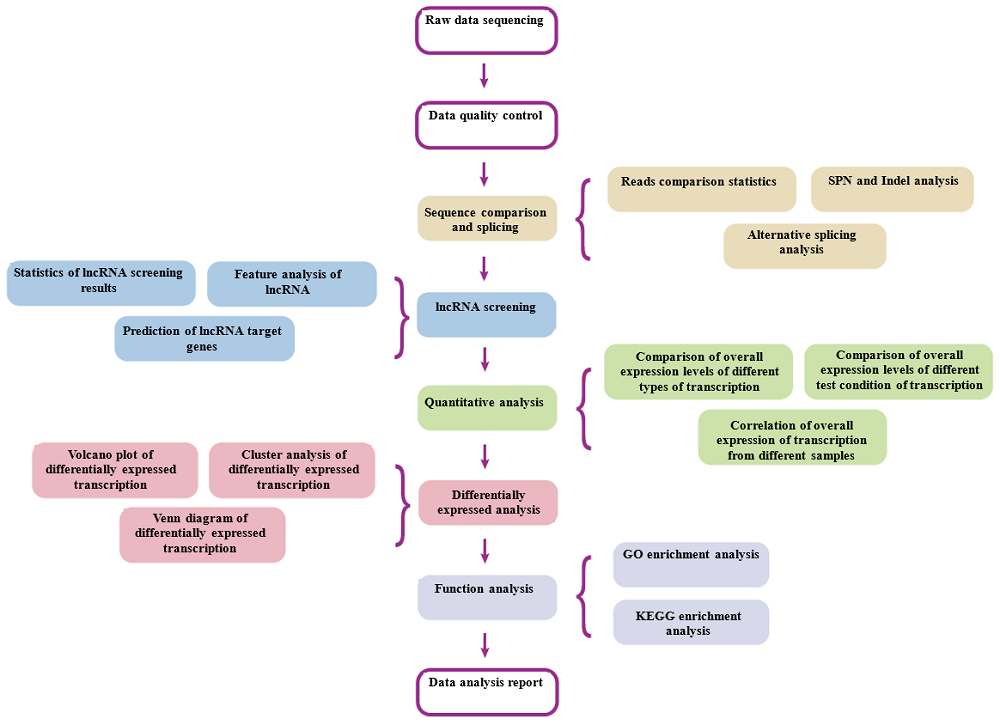
Analysis Workflow
-
Features
1. Data information of mRNA and lncRNA can be obtained simultaneously through one sequencing;2. Extensive experience with numerical samples being successfully sequenced.;
3. Powerful databases support, such as TCGA and HGMD;
4. Comprehensive analysis: It can be used for correlation analysis with other sequencing data, and the target relationship prediction analysis is comprehensive;
-
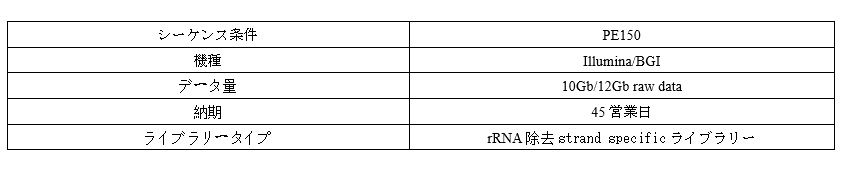
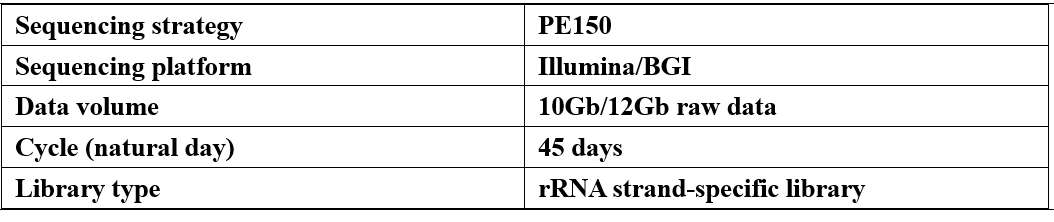
Parameter
-
Applications
Environmental threats, agronomic traits, medicine development, molecular markers development, growth and development, etc.
-
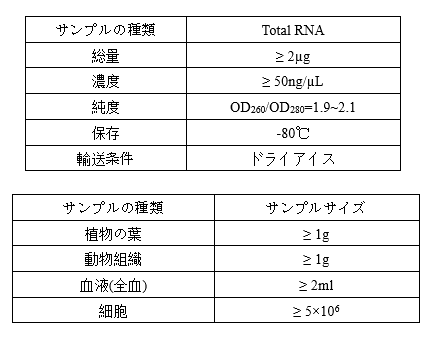
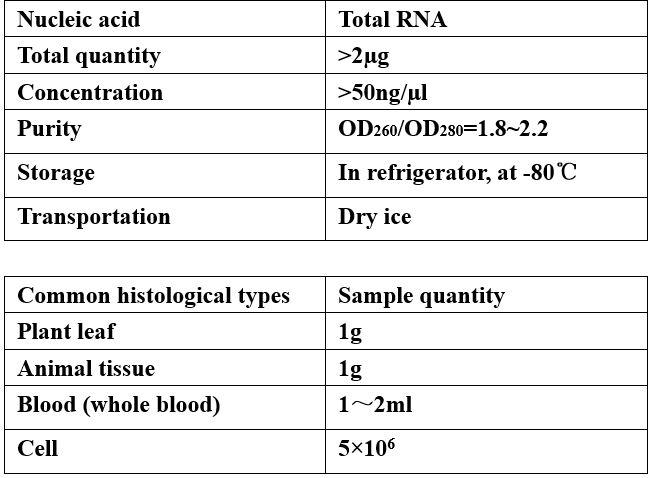
Requirements of Samples
-
Results Demo
Differentially expressed lncRNA volcano plot
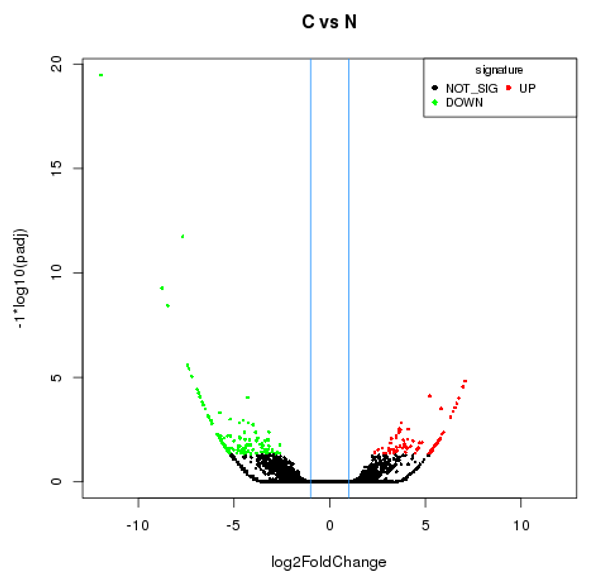
The input data of lncRNA differential expression is read count data obtained from lncRNA expression level analysis. For samples with biological replicates, we use DEseq for analysis; For samples without biological replicates, we use edgeR for analysis. Finally, we select genes with FDR (false discovery rate) less than 0.05 and fold change (FC) greater than or equal to 2 as the differentially expressed genes.
Cluster heat map
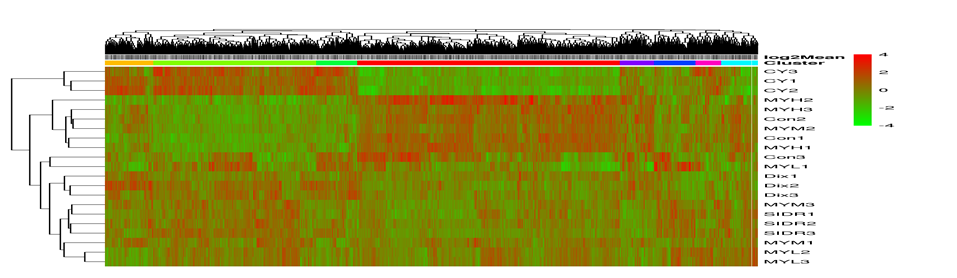
Cluster analysis is used to judge the expression patterns of differential genes under different experimental conditions; By grouping genes with the same or similar expression patterns into clusters, the functions of unknown genes or unknown functions of known genes can be identified; Because these similar genes may have similar functions or participate in the same metabolic process or cellular pathway.
GO enrichment
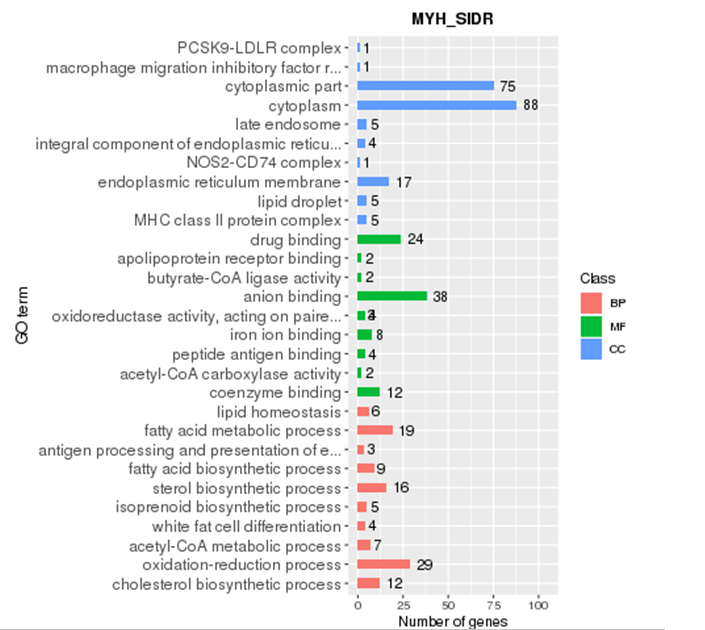
Gene Ontology (referred to as GO) is an international standard classification system for gene function. After screening the differential genes according to the experimental purpose, the distribution status of the differential genes in GO was studied in order to clarify the expression of the sample differences in the gene function in the experiment. The GO annotation information of most species is downloaded from Ensembl's Biomart database (http://asia.ensembl.org). If there is no GO annotation information for this species on Ensembl, the GO information annotated by yourself will be used.
KEGG enrichment
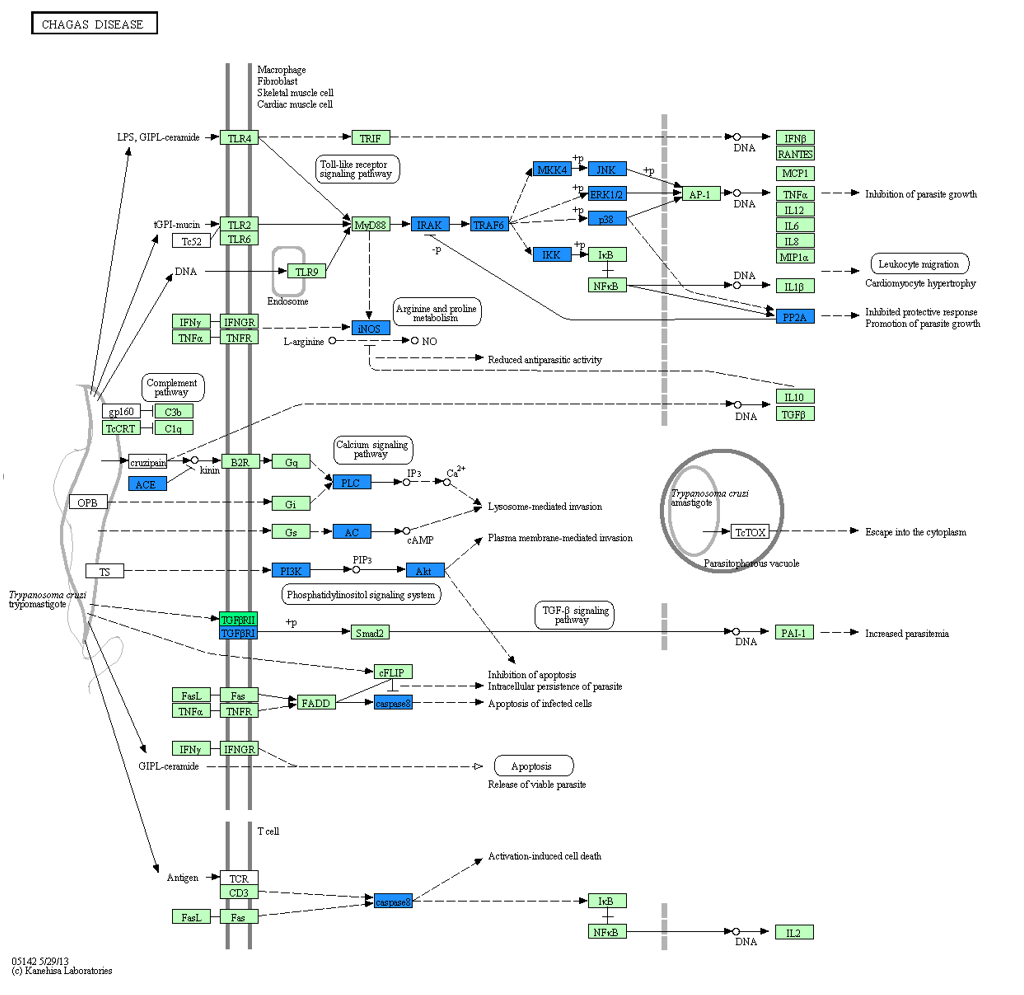
In organisms, different genes coordinate with each other to perform their biological functions. Through Pathway significant enrichment, the most important biochemical metabolic pathway and signal transduction pathway in which differentially expressed genes participate can be determined. KEGG (Kyoto Encyclopedia of Genes and Genomes) is the main public database for Pathway. Pathway significance enrichment analysis takes KEGG Pathway as a unit, and uses hypergeometric test to find Pathway that is significantly enriched in differentially expressed genes compared with the whole genome background.
PPI network analysis
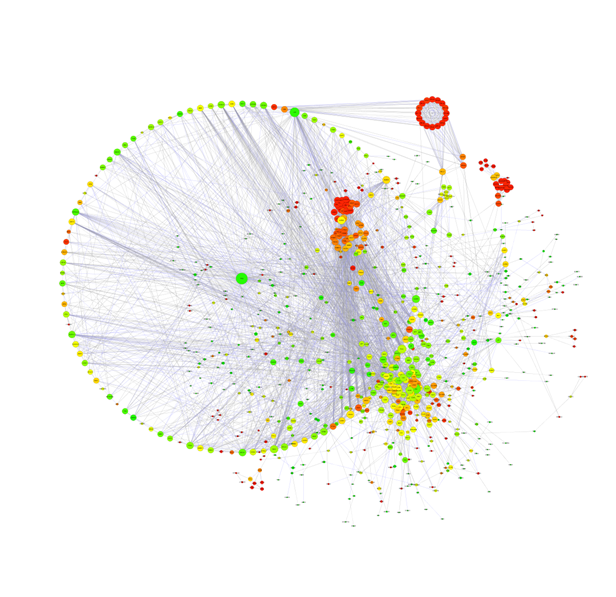
Use the interaction relationship in the STRING PPI database (http://string-db.org/) to directly extract the interaction relationship of the target gene set (such as the differential gene list) from the database for the species contained in the database to build a network; For species not included in the database, first, we use Blastx to compare the target gene set sequence to the protein sequence of the closely related or model species contained in the STRING database, and construct the interaction network by using the PPI relationships of the selected related or model species.


 Inquire
Inquire