-
- Q:
What types of samples can be extracted by miRcute miRNA extraction kits?
-
miRcute miRNA Isolation Kit (4992860) can extract miRNA from cells, tissues, serum plasma, plant tissues or total RNA containing miRNA. miRNAprep Pure FFPE Kit (4992863) can extract miRNA from FFPE samples. miRcute Serum/Plasma miRNA Isolation Kit (4992865) is specially designed for miRNA extraction from serum and plasma, while miRcute Plant miRNA Isolation Kit (4992733) is specially designed for miRNA extraction from plants, especially plants rich in polysaccharides and polyphenols.
- Q:
-
- Q:
What is the difference between the miRNA extracted by miRcute miRNA Isolation Kit and TRNzol reagent?
-
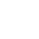
Using TRNzol to extract miRNA may result in selective loss of miRNA. Phenol and salt residues may exist in RNA extracted by TRNzol method, which has inhibitory effect on subsequent reverse transcription and other experiments might cause failure of subsequent experiments. Comparative data show that miRNA extracted by miRcute miRNA Isolation Kit can significantly reduce the residues of phenol and salt, while there is relatively high salt content in the total RNA extracted by TRNzol method (the absorbance value in the figure below represents the salt concentration).
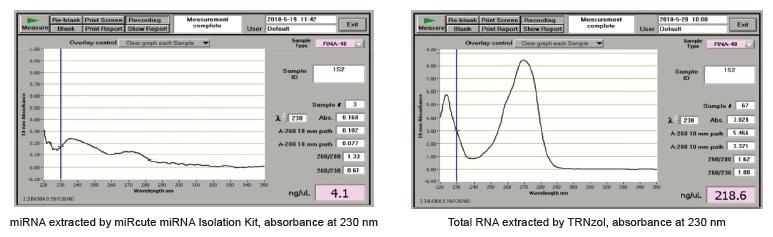
- Q:
-
- Q:
Is it suggested to measure the concentration of the miRNA extracted by miRcute miRNA extraction kits?
-
For tissues, cells and other samples, the electrophoresis detection of miRNA products extracted by miRcute miRNA Isolation Kit will show bright bands, but the products are mostly 5SRNA, 5.8SRNA and tRNA, so the absorbance value of this solution detected by UV spectrophotometer cannot truly represent the miRNA content in the products. For serum or plasma samples, electrophoresis is not recommended after miRNA extraction, because the circulating nucleic acid content is low, and no bands will show on an electrophoresis gel. The miRNA concentration detected by an ultraviolet spectrophotometer is also unreliable, but fluorescence quantitative PCR detection with higher sensitivity can be performed to detect miRNA concentration.
- Q:
-
- Q:
Can miRcute miRNA extraction kits be used to extract mRNA and miRNA respectively?
-
If mRNA and miRNA needs to be quantified separately, it is recommended to use the extraction solution of extracting total RNA (see the manuals for details). After conventional reverse transcription and poly(A)-tailing reverse transcription respectively, the target mRNA and miRNA are quantified respectively. If mRNA and miRNA needs to be extracted separately, it is suggested to use miRcute miRNA Isolation Kit for miRNA extraction. At the Column miRspin adsorption step, miRNA will be left in the filtrate, and the subsequent extraction can be carried out according to the steps in the manual. mRNA is bond to the Column miRspin, and after washing the miRspin with RNase-Free 70% ethanol (self-prepared) once, mRNA can be eluted by RNase-Free elution buffer or ddH2O.
- Q:
-
- Q:
What is the difference between miRNA reverse transcription by poly(A)-tailing and stem-loop method?
-
With the poly(A)-tailing, the reverse transcription product can be used for subsequent multi-miRNA detection, which is more accurate, convenient and fast, and does not need probe and stem-loop primers which requires high cost and long synthesis time. Although in principle, stem-loop method has higher specificity than poly(A)-tailing method, but with the progress of enzyme technology, there is no significant difference between the two methods.

- Q:
-
- Q:
How to avoid miRNA precursor amplification when the poly(A)-tailing miRcute Plus miRNA First-Strand cDNA Kit is combined with the miRcute Plus miRNA qPCR Kit (SYBR Green) to detect mature miRNA?
-
miRNA precursors present spatial hairpin structural characteristics (double-strand). The miRcute Plus miRNA First-Strand cDNA Kit with poly(A)-tailing adopts single-strand dependent poly(A) polymerase, and does not interact with double strand in the reaction with poly (A)-tailing, thus avoiding further reverse transcription and amplification of precursors. Moreover, the Quant RTase used subsequently is also a single-strand dependent reverse transcriptase, thus avoiding the amplification of precursors to the greatest extent.
- Q:
-
- Q:
What is the detection sensitivity of the miRcute Plus miRNA qPCR Kit (SYBR Green) combined with the poly(A)-tailing miRcute Plus miRNA First-Strand cDNA Kit?
-
The antibody-modified hot-start polymerase has excellent specificity and sensitivity. miRNA can be detected from total RNA as low as 5 pg (compared with stem ring method which can detect miRNA from 25 pg total RNA, the sensitivity is higher), and the difference of single base can be distinguished for miRNA family, so as to ensure high specificity of miRNA detection.
- Q:
-
- Q:
Can probe-based qPCR method be used for fluorescence quantification after reverse transcription with miRcute Plus miRNA First-Strand cDNA Kit (4992786/4992909)?
-
Yes. After reverse transcription, please use probe-based qPCR reagents (TIANGEN SuperReal PreMix Plus (Probe)/FastFire qPCR PreMix (Probe)/SuperReal PreMix Color (Probe)) and matching reverse primers to conduct probe-based miRNA quantitative detection. The detection probes shall be self-provided.
- Q:
-
- Q:
Can miRcute miRNA kits be used to detect piRNA, qiRNA and other microRNAs?
-
Yes. The miRcute miRNA extraction kit can extract all RNA with sizes less than 200 nt at the same time. These miRNAs can be reverse transcribed by miRcute Plus miRNA First-Strand cDNA Kit, and then quantified by SYBR method by using miRcute Plus miRNA qPCR Kit (SYBR Green). For designing and synthesizing forward quantitative primers, please consult TIANGEN customer service.
- Q: