Catalog Number|Packaging
Mat. No |
Ref. No |
No. of preps |
|
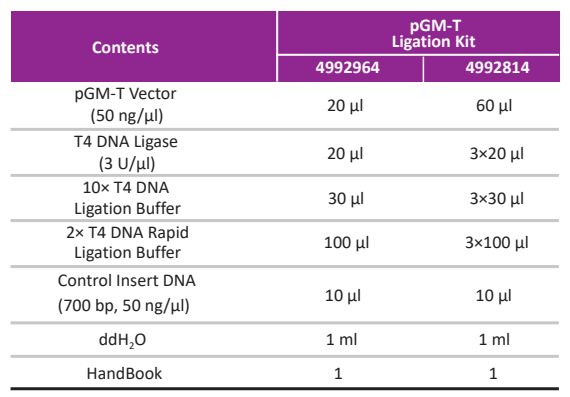
4992964 |
GVT202-01 | 10 μl×20 rxn |
|
4992814 |
GVT202-02 | 10 μl×60 rxn |
-
Description
The pGM-T Vectors are linearized vectors with a single 3´-terminal thymidine at both ends. The T-over hangs at the insertion site greatly improve the efficiency of ligation of PCR products by preventing
recircularization of the vector and providing a compatible overhang for PCR products generated by certain thermostable polymerases.
The pGM-T Vectors are high-copy-number vectors containing T7 and SP6 RNA polymerase promoters flanking a multiple cloning region within the α-peptide coding region of the enzyme β-galactosidase. Insertional inactivation of the α-peptide allows identification of recombinants by blue/white screening on indicator plates.
-
Kit Contents
-
Storage Condition
All reagents (containing vector and ligation reagents) are stored at -30~-15°C. Avoid repeated freezing and thawing. The shelf life is one year.
-
Important Notes
● It's necessary to use the control fragment during the transformation process, to confirm the reasons of the problems coming out in the experiment.
● It's suggested to leave part of ligation product. If the last step appeared problem, you could remedy quickly and don't have to repeat the ligation step.
● The bacterium volume adding on containing antibiotic SOB or LB solid culture agar could be adjusted according to the experiment. If the quantity of transformation DNA was large,the bacterium volume could be less than 100 μl; contrarily, the bacterium volume could be 200~300 μl. If it’s estimated that the clone was less, it could centrifuge 4,000 rpm for 2 min firstly, and then take out part of culture medium. Mix the remaining medium and bacterium, then extract suitable bacterium to add on containing antibiotic SOB or LB solid culture agar. Other remaining bacterium could store at 4°C. If the transformation bacterium colonies was less in the next day, it could extract stored bacterium to add on new containing antibiotic SOB or LB solid culture agar.
-
Protocol (All procedure must be done in asepsis environment)
This protocol is suitable for the cloning of PCR products with 3'-dA overhangs generated by Taq DNA polymerases or enzyme mixtures containg Taq DNA polymerare. (For blunt-end cloning, it is recommended to choose Lethal Based Fast Cloning Kit (Cat.no.4992815) or Lethal Based Simple Fast Cloning Kit (without MCS)(Cat.no.4992816)).
1. The vector must be thawed on the ice (avoid repeated freezing and thawing. Aliquot the vector before storage, and use appropriate amount for each time). Centrifugate the tube with vector shortly to remove drops from the inside of the lid.
2. Add all kinds of component in the asepsis tube as following: The mol ratio of vector and insert fragment must be controlled in 1:3~1:8. More fragments will disturb ligation reaction.
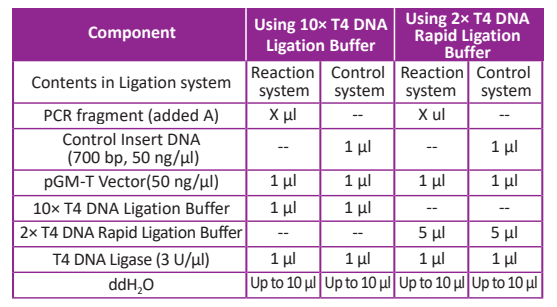
Note: There are two kinds of buffer in the kit, 10× T4 DNA Ligation Buffer and 2× T4 DNA Rapid Ligation Buffer. Don’t add two buffers in one tube.
3. Mix the solution in the tube gently, then centrifuges shortly. Incubate the tube with mixed solution at 22~26°C for 1~2 hours using 10× T4 DNA Ligation Buffer or incubate at 16°C overnight. If use 2× T4 DNA Rapid Ligation Buffer, incubate the tube with mixed solution at 22~26°C for 5~10 min. If it lasts for more than 15 min, the ligation efficiency would reduce. After reaction, place the tube on the ice.
Note: It’s suggested that incubate the tube with mixed solution at 16°C overnight using 10× T4 DNA Ligation Buffer. pGM-T Ligation Kit Handbook Using 10× T4 DNA Ligation Buffer.
4. Transformation
a. Prepare agar plates for transformation.
Add 16 µl IPTG (50 mg/ml) and 40 µl X-Gal (20 mg/ml) on agar plate surface which contain corresponding antibiotic. Smear completely using a sterile bent glass rod or a specialized spreader. Then put that plate at 37°C for 1~3 hours with no light.
b. Transformation step
i. Remove tube(s) of TOP10 competent cells from storage and placein an ice bath until just thawed. Carefully add part of ligation-reaction mixture to 50~100 µl TOP10 competent cells. The adding
volume of ligation-reaction mixture should be less than one tenth of competent cell volume. Gently flick the tubes to mix and place them on ice for 30 min. (If necessary, use control plasmid pUC19
to transform competent cell to detect transformation efficiency. Add 0.1 ng pUC19 to another tube with proper competent cell, and then, other steps go along with the step of transformation of ligation product during the same period.)
ii. Heat-shock the cells for 90 sec in a water bath at exactly 42°C (do not shake). Immediately return the tubes to ice for 2~3 min (do not shake).
iii. Add 250~500 μl SOC or LB culture medium preheated to 37°C per tube (not containing antibiotic), and then incubate for 45 min at 37°C with shaking (~150 rpm).
iv. Mix bacterium in the tube completely. Then plate 100 μl transformation culture onto each SOB or LB agar plate containing antibiotic to ensure good separation of colonies for subsequent single-colony isolation. Smear bacterium completely with asepsis elbow glass stick. After the surface of plate is dry, then put the plate at 37°C for 12-16 hours.
5. Detection
a. General detection: pipet the transformation mixture into 1~5 ml liquid LB culture medium(containing 50~100 μg/ml ampicillin), and culture at 37°C overnight with shaking. Save bacterium strain and
extract plasmid. To detect whether the fragment has inserted rightly using PCR or restriction enzyme digestion.
b. Quick detection: to detect whether the fragment has inserted rightly using bacterium PCR directly.c. Sequencing: sequence the fragment after general or quick detection.
c. Sequencing: sequence the fragment after general or quick detection.
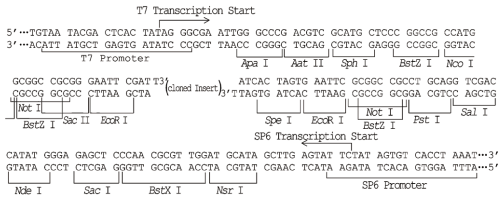
pGM-T Vector Map
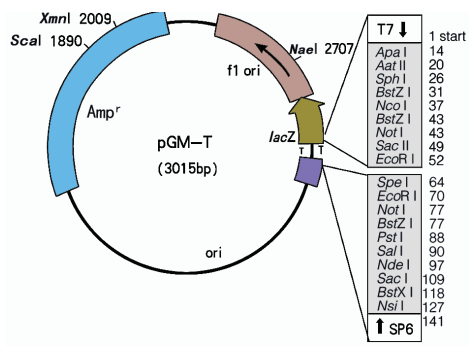
pGM-T Vector Cloning Site
-
Sort by
-
Date
Date(
)
Date
Date(
)
Impact Factor
IF(
)
Impact Factor
IF(
)



 Inquire
Inquire