Catalog Number|Packaging
Mat. No |
Ref. No |
No. of preps |
|
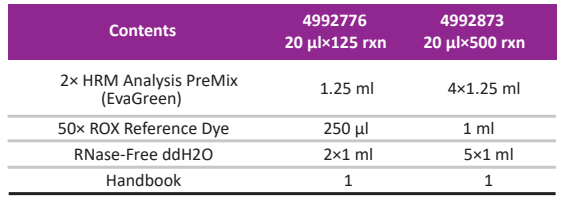
4992873 |
GFP210-02 | 20 μl×500 |
|
4992776 |
GFP210-01 | 20 μl×125 |
-
Features
● HRM Analysis PreMix uses antibody modified hot start DNA polymerase, along with the specially optimized buffer solution, provides a high sensitive and high efficient analysis method with wide sample range.
● HRM Mix includes EvaGreen saturation dye, which has high melting resolution under saturation condition, so it could detect the mutation of single base.
● The unique buffer system in this kit strengthens the stability of melting curve, increasing the specificity of amplification.
● This Kit includes ROX Reference Dye, which can eliminate the interference of signal background and revise the deviation of fluorescence signal between different wells.
-
Description
HRM analysis Kit (EvaGreen) combines the advantages of saturation dye EvaGreen and antibody modified enzyme, and is specially designed to perform High Resolution Melting (HRM) analysis. A 2×pre-mixed solution is included in this kit which greatly simplifies the preparation of qPCR reaction mixture. It is highly sensitive and could be used to analyze different types of templates.
Unlike SYBR Green, high concentration of EvaGreen dye will not inhibit PCR reaction, but could increase dsDNA products binding, so it is called "saturation dye". In contrast with SYBR Green, "dye rearrangement" will not happen when using EvaGreen, so it could properly distinguish the difference on single base of amplification products.
This Kit can be used in known SNP analysis, unknown mutant gene scan and Methylation Specific PCR (MS-PCR).
-
Kit Contents
-
Applications
● Known SNP typing.
● Unknown SNP screening.
● Unknown mutant genes scanning.
● Methylation PCR analysis.
-
Storage Condition
The HRM Analysis Kit (EvaGreen) can be stored at -30~-15°C for one year. It should be stored immediately after receipt at -30~-15°C, protected from light. Thaw the 2× HRM Analysis PreMix and 50× ROX Reference Dye and mix thoroughly before use. If the 2× HRM Analysis PreMix and 50× ROX Reference Dye are thawed but not used, it is important to thoroughly mix them prior to refreezing. The delamination of salts during the thawing process and subsequent crystallization during freezing will damage the enzyme and decrease product performance. For frequent use, HRM Analysis PreMix can be stored at 2-8°C for 3 months. Repeated freeze-thaw cycles should be avoided.
-
Important Notes
● This Kit includes the fluorescent dye EvaGreen. Store the reagent in dark and avoid direct exposure to strong light during the preparation of PCR reaction mixtures.
● Performance of reagents may reduce without mixing well. Please gently mix the reagents by inverting the tubes and centrifuge briefly prior to use. Do NOT vortex and avoid producing bubbles.
● The purity of primers is important for the specificity of PCR. Primers purified by PAGE or more superior methods are recommended.
● In a 20 µl reaction volume, the amount of genomic DNA template is usually less than 100 ng. Please use the same template amount between different reactions. Template purity should be: OD260/280 1.6-2.0, OD260/230 1.5-2.0.
● Since HRM have extreme high sensitivity, so 50 µl reaction volume is recommended. Large reaction volume could increase the experimental repeatability, and reduce the negative effect of experimental error to melting curve.
● Primer design: Shorter PCR product could increase the HRM resolution. So beside the common principles of primer design, please make the length of PCR product within 80-120 bp. SNP site should be designed in the middle of PCR product sequence.
-
Principle of Operation
SYBR Green is the most widely used fluorescent dye in Real Time PCR, but its concentration need to be very low in the PCR reaction mixture since it has inhibition effect toward the PCR reaction. The binding condition of SYBR Green and dsDNA is not saturated, so it is called "unsaturation dye". This trait of SYBR Green will lead to the "dye rearrangement" effect, and then influence the resolution of melting curve. In this condition the difference between single bases cannot be distinguished by analyzing melting curve when using SYBR Green as fluorescent dye.
EvaGreen is a novel fluorescent dye which can be used both in Real Time PCR and HRM analysis. This dye can selectively bind to dsDNA through a specific mechanism called "release according to requirement". This mechanism not only maximally reduces the PCR inhibition effect of fluorescent dye, but also allows the HRM analysis to detect single base differences when the fluorescent dye is under saturation condition.
-
Protocol
● Set up the HRM PCR reaction system
Note: 2× HRM Analysis PreMix and 50× ROX Reference Dye should be stored protected from light.
● Thaw 2× HRM Analysis PreMix (if stored at -30~-15°C) , 50× ROX Reference Dye, template, primers and RNase-Free ddH2O. Completely mix and equilibrate reagents to room temperature before use.
● All the HRM PCR reaction steps should be operated on ice.
Reaction system
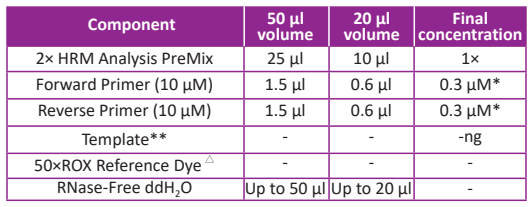
Regular reaction volume is 20 µl, but please use 50 µl reaction volume if possible. Large reaction volume could reduce the negative effect of experimental error to melting curve.
*A final primer concentration of 0.3 μM is optimal for most applications. Higher primer concentration can be used when the amplification efficiency is not favorable. If non-specific amplification is observed, the primer concentration should be reduced. For further optimization, a primer titration of 0.2-0.5 μM can be performed.
**Since the HRM analysis is extremely sensitive, please use the same template amount between different reactions.
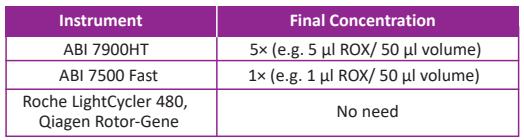
△The optimal concentration of ROX Reference Dye for commonly used Real-Time PCR instruments:
● Real-Time Amplification
Typically, two-step PCR is recommended. However, if two-step PCR does not yield favorable results (e.g. non-specific amplification caused by low template concentration or reduced amplification efficiency induced by low Tm value) three-step PCR is recommended.
Two-step PCR:
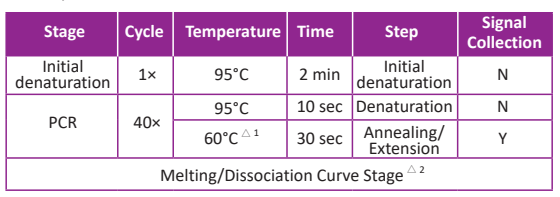
Three-step PCR
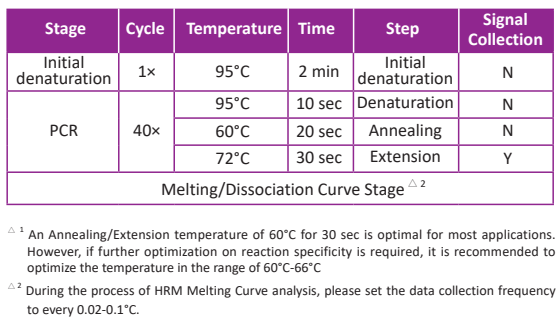
● Close the tubes and mix samples gently. Brief centrifugation can be performed to collect residual liquid from the walls of the tubes.
● Place the PCR tubes in the thermal cycler and then start the PCR cycle.
-
Case Study
Retrieve known SNP from NCBI database for analysis, the information of SNP is as follows:
RefSNP number: rs174538
Gene Name: FEN1 (flap structure-specific endonuclease 1)
Sequence: CGCTCCGCCACCGGAAGAACACGTCG[A/G]CAGGAGCAGGCGCCTAGCACAACCG
Allele Frequency: A=0.314, G=0.686
Design the primer according to the sequence of the both sides of SNP sites:
FEN1_F: CCTCAACGCTCTCACCATTTTG;
FEN1_R: GGCACTTCCTTTTCCGGTTGTG;
The length of amplified PCR product is 108 bp.
Setup the PCR reaction, run the PCR program on Roche LightCycler480, 24 samples are analyzed.
Reaction system:
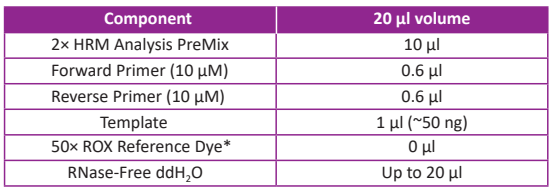
*ROX Reference Dye is not required on LightCycler 480
Reaction Program:
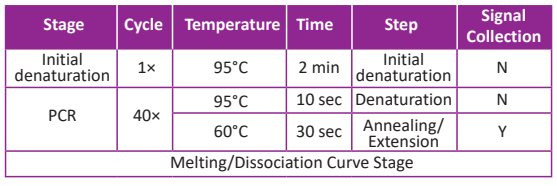
Experimental Example
Genomic DNA was extracted from 100 μl of human blood sample using TIANamp Blood DNA Kit. 50 ng DNA was loaded for the real time PCR detection. According to NCBI SNP library, the known SNP of FEN1 gene (RS 174538) is selected and detected using Roche LightCycle480. The results are as follows:
The results show that HRM Analysis Kit is an ideal SNP locus analysis method with high repeatability of melting curve of the same genotype, clear resolution of different genotypes and no misjudgment.
SNP information: ......CGGAAGAACACGTCG[A/G]CAGGAGCAGGCGCCT…… (Allele Frequency: A=0.314, G=0.686)
Primer: For 108 bp fragment (FEN1_F: CCTCAACGCTCTCACCATTTTG; FEN1_R: GGCACTTCCTTTTCCGGTTGTG)
-
Sort by
-
Date
Date(
)
Date
Date(
)
Impact Factor
IF(
)
Impact Factor
IF(
)



 Inquire
Inquire