Catalog Number|Packaging
Mat. No |
Ref. No |
No. of preps |
-
Description
The regulatory network of noncoding RNA of 18-40nt in length, as an important regulatory factor, play an important role in physiological processes such as gene expression regulation, biological ontogeny, metabolism and disease occurrence. All Small RNAs in cells or tissues can be sequenced deeply and analyzed quantitatively by high-throughput sequencing technology.
-
Technical Workflow
Scheme design → Sample processing → Library construction sequencing → Data analysis → After-sales service
-
Analysis Workflow
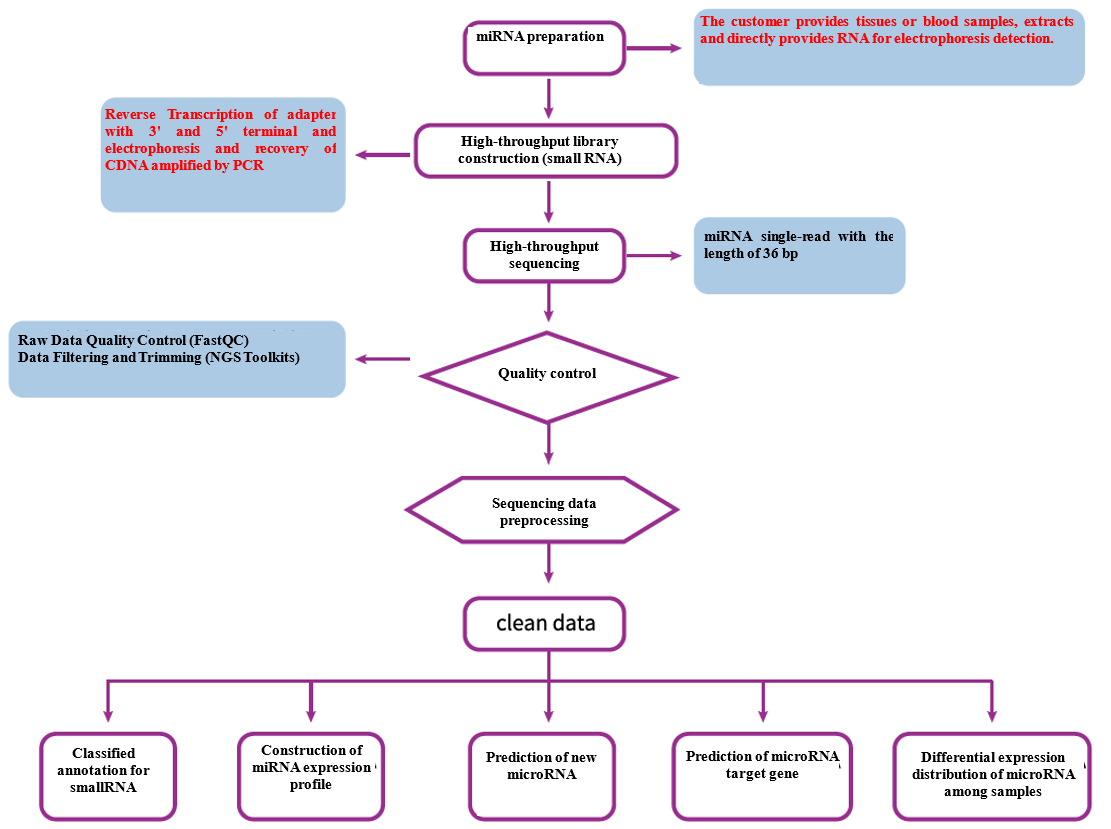
-
Features
1. 30-day full cycle;
2. Senior technical teams, with more than ten years of information analysis experience, can provide a variety of personalized analysis;
3. Each step should be of high quality and accuracy from extraction to library construction, and sequencing to analysis.
4. Free correlation analysis for both small RNA and mRNA expression levels to investigate the regulatory networks.
-
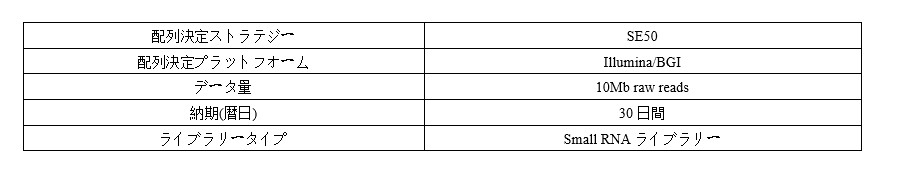
Parameter

-
Applications
Environmental threats, agronomic traits, medicine development, molecular markers development, growth and development, etc.
-
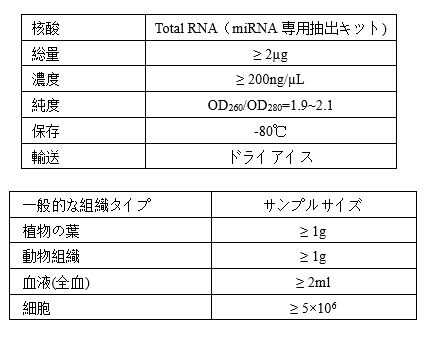
Requirements of Samples
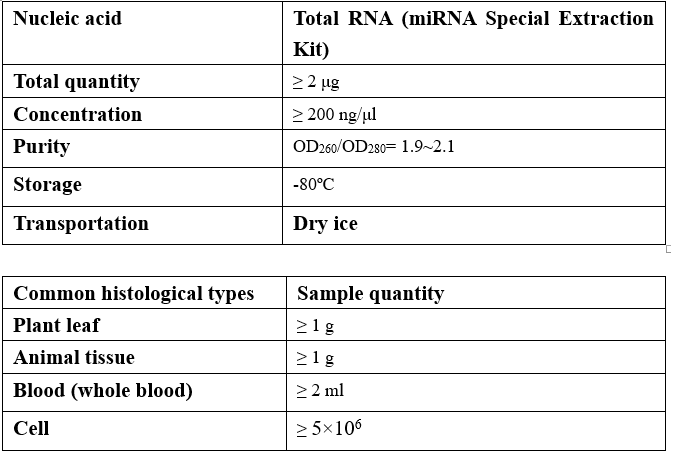
-
Results Demo
Differential miRNA volcanic plot
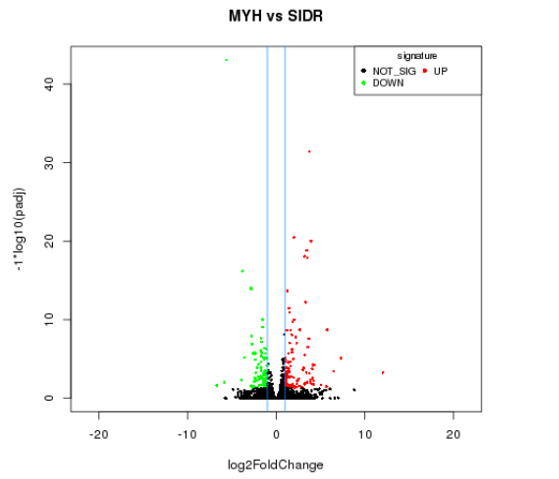
The input data of miRNA differential expression is read count data obtained from miRNA expression level analysis. For samples with biological replicates, we use DEseq for analysis; For samples without biological replicates, we use edgeR for analysis. Finally, we select genes with FDR (false discovery rate) less than 0.05 and fold change (FC) greater than or equal to 2 as the differentially expressed genes.
Cluster heat map
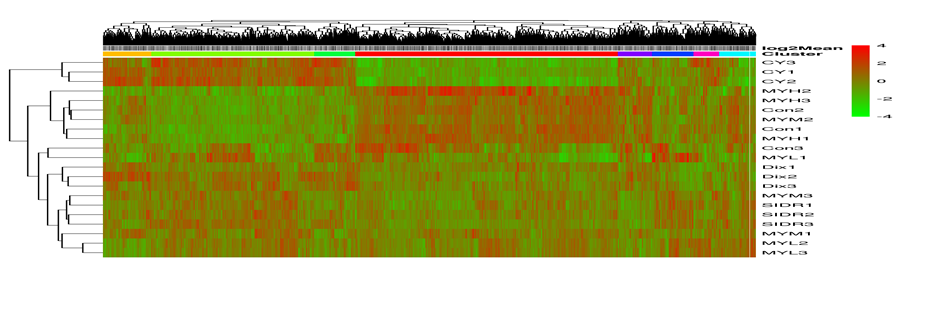
Cluster analysis is used to judge the expression patterns of differential miRNA under different experimental conditions; By clustering miRNAs with the same or similar expression patterns, the function of unknown and known miRNAs can be judged; Because these similar miRNAs may have similar functions or participate in the same metabolic process or cellular pathway.
GO enrichment of target gene

Gene Ontology (referred to as GO) is an international standard classification system for gene function. After screening the differential genes according to the experimental purpose, the distribution status of the differential genes in GO was studied in order to clarify the expression of the sample differences in the gene function in the experiment. The GO annotation information of most species is downloaded from Ensembl (http://asia.ensembl.org). If there is no GO annotation information for this species on Ensembl, the GO information annotated by yourself will be used.
-
Sort by
-
Date
Date(
)
Date
Date(
)
Impact Factor
IF(
)
Impact Factor
IF(
)



 Inquire
Inquire