Catalog Number|Packaging
Mat. No |
Ref. No |
No. of preps |
|
4992910 |
GKR107-01 | 20 µl×25 rxn |
|
4992911 |
GKR107-02 | 20 µl×100 rxn |
-
Features
● High enzyme activity and efficiency: High reverse transcriptase activity and good compatibility in subsequent experiments.
● Wide substrate range: Suitable for all RNA, especially RNA templates with complex secondary structures.
● Long RT length: The synthesis of the cDNA first strand can reach 12 kb.
● Simple operation: Simply add the required components in one step without adding any reagent during the operation.
-
Description
TIANScript II RT Kit supplies all the required reagents for first-strand cDNA synthesis, including the TIANScript II RTase which is modified from M-MLV, with higher affinity to template and lower RNase H activity. Therefore it has the ability to read complex secondary structure of template and reverse transcribe long RNA in the process of first-strand cDNA synthesis. The 5× TIANScript II RTase Buffer in the kit not only guarantees high enzyme activity of new RTase, but also expands the template input range. cDNA synthesized by this kit has high quality and is good for downstream analysis.
-
Kit Contents
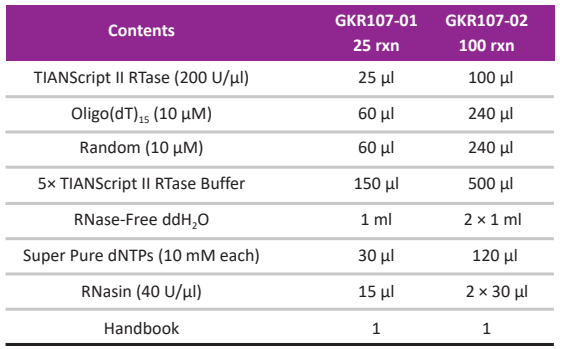
-
Applications
● Synthesis of first-strand cDNA.
● Construction of cDNA library.
● Two step RT-PCR.
● RACE analysis.
-
Storage Condition
All the components can be stored at -30~-15°C for up to 12 months. -
Source of Enzyme
Recombinant E. coli which contains improved M-MLV gene. -
Important Notes
● Solutions that used for cDNA synthesis should be treated with DEPC and autoclaved before use. For reagents not suitable for autoclaving, prepare the reagents with the sterilized container and water, and then filter to obtain the final solutions.
● Avoid genomic DNA contamination in RNA sample.
● Repeated freezing and thawing of RNA should be avoided. Keep the dissolved RNA on ice.
● All the components of the kit should be stored at -30~-15°C.
● When using Random Primers, please note the ratio of the amount of primers and the amount of RNA. It is suggested to use 50 ng of Random Primers for every 5 µg total RNA template. Increase the ratio of Random/RNA will improve the synthesis of short cDNA (~500 bp); and decrease the ratio of Random/RNA will improve the synthesis of long cDNA.
● For RNA templates rich in secondary structures, it is suggested to heat at 65°C for 5 min before the reaction.
● If the reverse primer of PCR reaction would be used in reverse transcription, in order to avoid nonspecific amplification led by primer mismatch, the whole transcription process could be done at 50°C.
● We suggest to terminate the synthesis at 70°C for 15 min in order to avoid cDNA damage.
● When using the enzymes, please mix enzymes gently to avoid bubbles; please centrifuge carefully to collect the enzyme before pipetting, and please aspirate slowly since enzymes are sticky.
Note: For long cDNA synthesis, please prepare fresh, integrate and pure RNA template.
-
Unit Definition
● 1 unit (U) is defined as the amount of enzyme required to catalyze the incorporation of 1 nM of dNTP into acid-insoluble in 10 min at 37°C, using polyA.poly(dT)12-18 as template and primers.
-
Protocol
● Thaw the template RNA on ice; Thaw the primers, 5× TIANScript II RTase Buffer, Super Pure dNTPs and RNase-Free ddH2O at room temperature (15-30°C) and then place all the reagents on ice. Mix all the reagents by vortex and briefly centrifuge before use to collect drops from the inside of the tube to the bottom.
● Set up the reaction in a nuclease-free microcentrifuge tube on ice according to the following table:
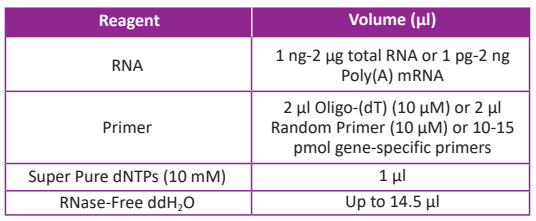
● Incubate at 65°C for 5 min and then cool on ice immediately for 2 min.
● Continue to setup the reaction according to the following table:
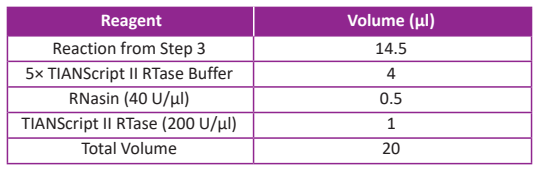
● If using Random Primers, incubate the microcentrifuge tube at 25°C for 10 min. If using Oligo-dT or gene-specific primers, skip this step.
● Incubate at 42°C for 60 min.
● Heat the reaction mixture at 85°C for 5 min (or 70°C for 15 min) to terminate the reaction, and then place on ice for downstream experiments or store at -20°C immediately.
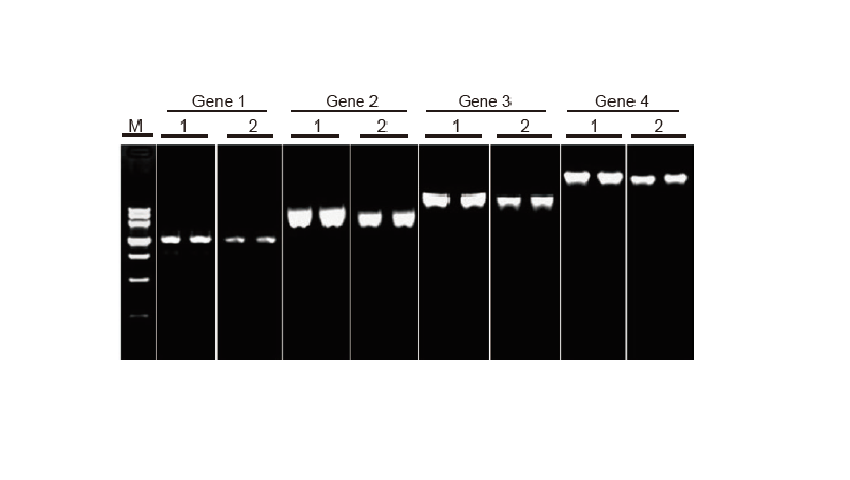
Experimental Example
Reverse transcription ability of TIANScript II RT Kit for different length fragments
Method: Reverse transcription: Refer to the instruction manual of TIANScript II RT Kit.
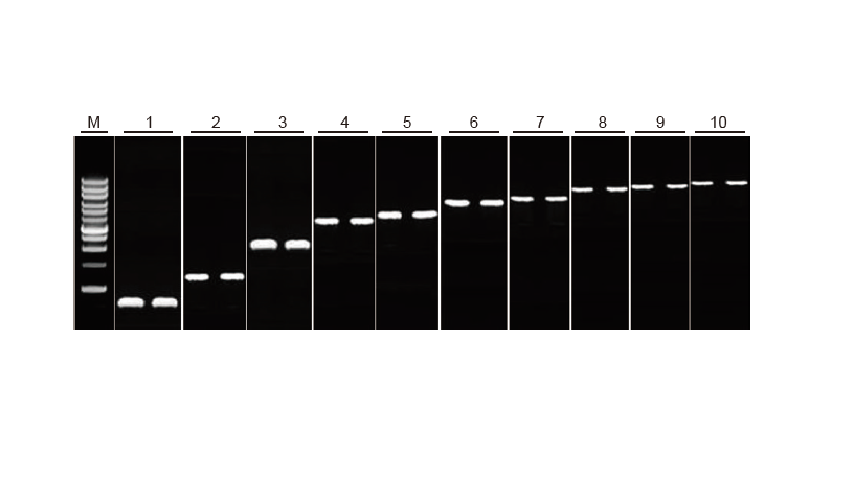
Results: The gel picture shows the amplification results of 10 target genes with different lengths after reverse transcription of 1 μg of total RNA. 2 μl of reverse transcription products were loaded per lane.
Amplification system (PCR): 20 μl; Sample load: 5 μl;
Marker: D15000+1 kb DNA Ladder; Gel concentration: 1%;
Electrophoresis conditions: 6 V/cm, 20 min
Diagram of each lane: M: DNA Marker; 1: Product length: 120 bp; 2: Product length: 1 kb; 3: Product length: 2.5 kb; 4: Product length: 3.2 kb; 5: Product
length: 4.6 kb; 6: Product length: 6.8 kb; 7: Product length: 7.6 kb; 8: Product length: 8.9 kb; 9: Product length: 10 kb; 10: Product length: 12 kb;
-
Sort by
-
Date
Date(
)
Date
Date(
)
Impact Factor
IF(
)
Impact Factor
IF(
)



 Inquire
Inquire