Catalog Number|Packaging
Mat. No |
Ref. No |
No. of preps |
|
4992261 |
GNG102-01 | 24 rxn |
|
4992262 |
GNG102-02 | 96 rxn |
-
Features
● The one-tube enzymatic reaction completes the end repair of double-stranded DNA fragments and dA-tailing reaction in one step.
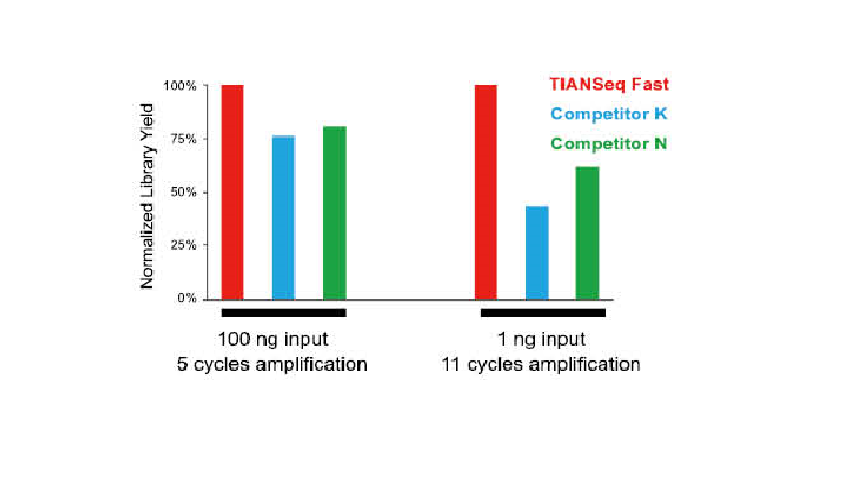
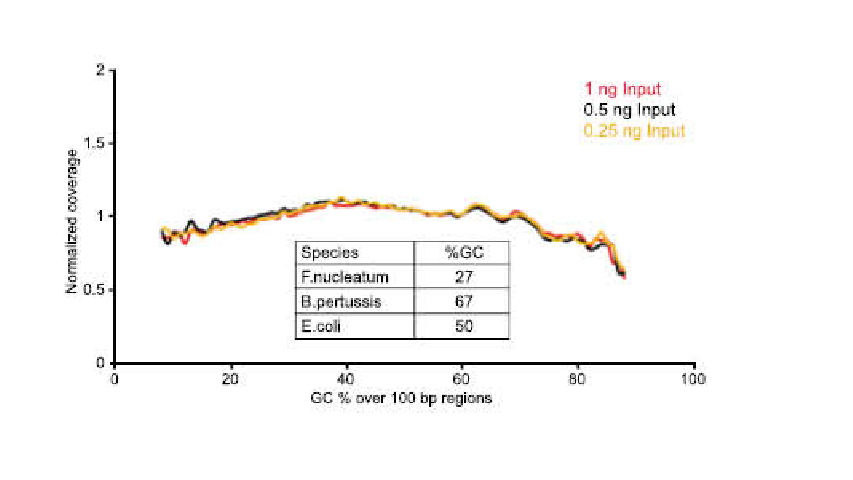
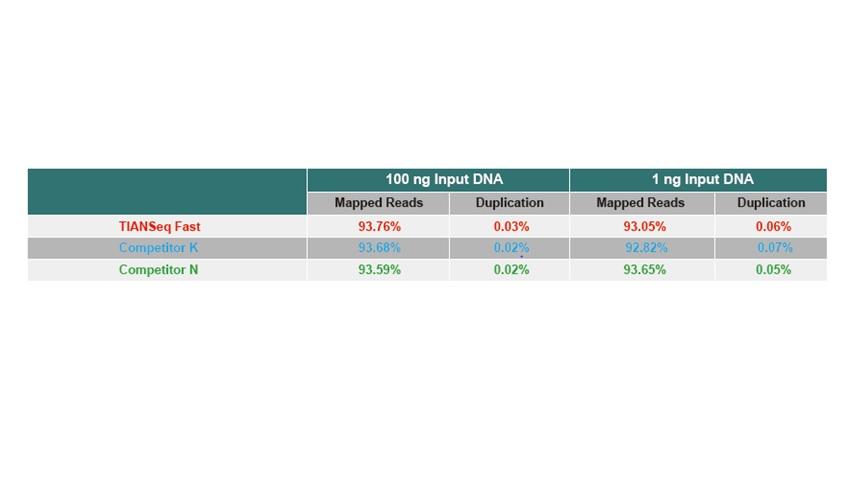
● There is no base bias in PCR enrichment process with uniform sequence coverage.
● High conversion efficiency of the library, and the input of DNA sample can be as low as 0.25 ng.
-
Description
The TIANSeq Fast DNA Library Kit (illumina) is a DNA library construction kit specially optimized for illumina high-throughput sequencing Platforms. This product combines the end-repair and 3'-end dA-tailing reactions in one tube. This kit can be applied for short DNA fragments or pre-sheared double-stranded DNA which has been fragmented by sonication, chemical treatment or enzyme-based reaction. Meanwhile, the obtained product can be directly used for adaptor ligation without additional purification step. In addition, the PCR amplification reagent in this kit has been specially optimized to ensure high yield, good fidelity and no base preference for the amplified DNA library. The one-step streamline workflow of this kit skips multiple purification steps, and make sure the entire library construction process can be performed in 2.5 hr. This kit can achieve high library construction efficiency even for low input DNA samples.
-
Kit Contents
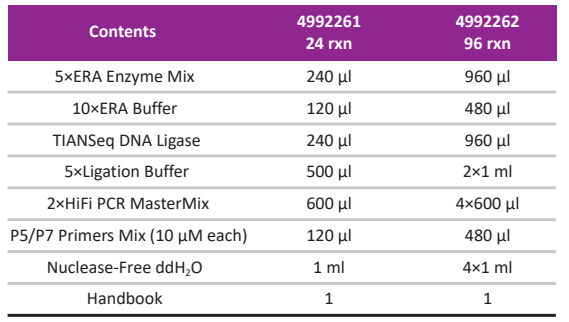
-
Applications
Ideal choice for the DNA library construction of Illumina high-throughput sequencing Platforms. -
Sample Input
0.25 ng~1 μg DNA. -
Required Reagents
● TIANSeq Single-Indexed Adapter (Illumina) (Cat# 4992641/4992642/4992378)
● TIANSeq Size Selection DNA Beads (Cat# 4992358/4992359/4992979)
-
Storage Condition
The kit should be stored at -30~-15°C. Avoid repeated freezing and thawing. The shelf life is one year. -
Product Highlights
● Enzyme-based reaction in a single tube. End-repair of double-stranded DNA fragments and dA-tailing reaction can be achieved in one step.
● No base preference in the process of PCR amplification, and the sequencing uniformity is ensured.
● High library construction efficiency can be achieved with the DNA input as low as 0.25 ng.
-
Precautions
● Attention should be paid in the operating process to avoid cross-contamination between nucleic acid samples and products.
● Please use RNase- or DNase-free pipette tips or EP tubes for the experiment.
● Before starting, wipe down work area with RNase and DNase cleaning reagents such as RNase Away (Molecular BioProducts, Inc). Make sure there is no contamination of RNAse and DNase.
● Before the library amplification, make sure the thermal cycler is calibrated and in a stable state.
● Please read the protocol carefully before the experiment. If test suspension is needed or the downstream test is not needed to be carried out immediately, the test products can be frozen and stored at -20°C and the subsequent test can be planned accordingly.
-
Protocol
● DNA Fragmentation
This kit does not contain DNA fragmentation related reagents. For the fragmentation process of DNA, customers can choose among the common methods such as ultrasonic treatment, chemical treatment and enzyme treatment. Please refer to the relevant product description for detailed operation.
● End repair/dA-tailing
● Preparation:
● Before the experiment, it is critical to determine the concentration of the input DNA and the buffer the DNA is dissolved in.
Note: It is critical to determine the quality and concentration of the input DNA sample, especially when the input amount is below 100 ng. Qubit, Picogreen or other Fluorometric methods are recommended for accurately quantifying the DNA. In addition, please confirm the buffer in which the DNA is dissolved, as the protocol needs to be slightly adjusted with different buffers.
● Place the reagents on ice. Once thawed, mix the 5× ERA Enzyme Mix by finger flicking (do not vortex to mix). The remaining reagents can be mixed by quick vortexting.
● Procedures
● If DNA is dissolved in deionized water, 10 mM Tris, Buffer EB or 0.1× TE, set up the end repair/dA-tailing reaction following the steps below.
● Prepare the following program (table below) into a thermal cycler. Turn on the hot lid and set the temperature to 70°C.
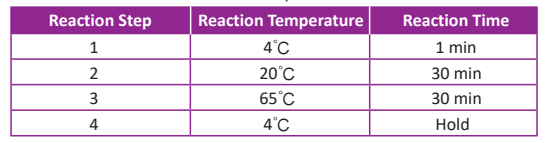
● Prepare the reaction mix on ice according to the following table in a new 200 µl thin-walled tube. Mix well by gently pipetting. Do not vortex to mix.
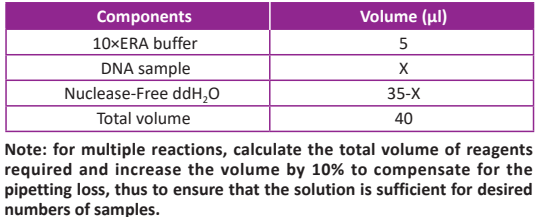
● Add 10 μl 5× ERA Enzyme Mix to the thin-walled tube from Step (2), and gently mix by pipetting up and down for 10 times. Do not vortex to mix.
Note: This step needs to be operated on ice the whole time.
● Pulse-spin the thin-walled tube, and immediately put it in the thermal cycler that has been pre-cooled to 4°C, then start the cycling program.
● When the cycling program is completed, remove the thin-walled tube from block and put it on ice.
● Proceed to the adapter ligation step immediately.
● If DNA is dissolved in other solutions, determine the concentration of saline ions in the solution, especially EDTA. EDTA has an apparent influence on the reaction. If the concentration of EDTA in the solution is uncertain or in a high level, we recommend applying TIANSeq Size Selection DNA Beads (Cat# 4992358/4992359/4992979) for the purification of DNA. The purification steps are as follows:
● Equilibrate the beads at room temperature for 20 min.
● If the volume of DNA solution is less than 50 μl, adjust the volume to 50 μl with nuclease-free deionized water.
● Add 1.8× (90 μl) of thoroughly vortexed the magnetic beads to DNA solution and mix well by pipetting up and down for 10 times. If the volume of DNA solution is greater than 50 μl, scale the volume of the completely vortexed the magnetic beads appropriately such that the ratio of beads to DNA is 1.8×.
● After incubating at room temperature for 5 min, place the reaction tube on the magnetic stand for 5 min to pellet the magnetic beads, and carefully remove the supernatant.
● Place the tube on the magnetic stand and add 200-500 μl freshly prepared 80% ethanol (the ethanol should be just enough to immerse all the beads) to the reaction tube, then gently pipette up and down for 3-5 times to wash the magnetic beads (do not disturb the beads). Pellet the magnetic beads with a magnetic stand for 30 sec and discard the supernatant.
● Place the reaction tube on the magnetic stand, open the centrifuge tube cover and air-dry the beads at room temperature for 5-10 min until the magnetic beads are dried.
Note: Do not over-dry the beads, or it will cause the decrease of the yield.
● Remove the reaction tube from the magnetic stand, and thoroughly resuspend the dried beads in 32.5 μl 10 mM Tris-HCl (pH8.0) by pipetting up and down for 10 times. Place the tube at room
temperature for 5 min, and pellet the beads on the magnetic stand for 5 min. Carefully transfer 30 μl of supernatant into a new centrifuge tube after the magnetic beads are attached.
● Use Quibit, Picogreen or other Fluorometric methods to determinethe concentration of purified DNA.
● Adapter Ligation
Place the reagents on ice. Once thawed, mix the TIANSeq DNA Ligase by gentle finger flicking (do not vortex to mix). The remaining reagents can be mixed by quick vortexing.
● After the end repair/dA-tailing reaction, add Y μl adapter solution to the 50 μl reaction, mix gently by pipetting and put on ice.
Notes: This kit does not contain the DNA adapter for sequencing.Please refer to the usage conditions provided by the adapter supplier. TIANSeq Single-Indexed Adapter (Illumina) (Cat# 4992641/4992642/4992378) is recommended. To achieve higher ligation efficiency, we recommend the molar ratio of the adapter to the DNA fragments in the reaction mix to be between 10:1 to 200:1.
● Prepare the reaction master mix according to the table below. Mix gently by pipetting and then keep it on ice.
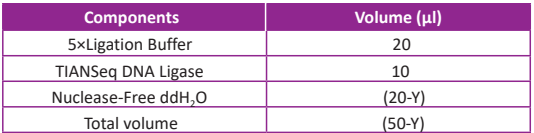
● Add the prepared (50-Y) μl ligation master mix to the reaction solution prepared in Step 1 to generate a 100 µl reaction mix, then gently mix by pipetting up and down for 10 times. Incubate the ligation reaction at 20°C for 15 min.
Notes: If this step is performed using a thermal cycler, turn on the hot lid and set the temperature ≤ 40°C.
● It is recommended to use 1× (100 μl) TIANSeq Size Selection DNA Beads (4992358/4992359/4992979) for the purification of ligation products.
The steps are as follows:
● Equilibrate the magnetic beads at room temperature for 20 min.
● Vortex the magnetic beads to fully suspension. Add 100 μl of the thoroughly vortexed the magnetic beads to the solution in Step 3, and mix well by pipetting up and down for 10 times.
● Incubate the mix for 5 min at room temperature. Place the reaction tube on the magnetic stand for 5 min. After the magnetic beads are completely attached, carefully remove the supernatant.
● Place the tube on the magnetic stand and add 200-500 μl freshly prepared 80% ethanol (the ethanol should be just enough to immerse all the beads) to the reaction tube, then gently pipette up and down for 3-5 times to wash the magnetic beads (do not disturb the beads). Pellet the magnetic beads with a magnetic stand for 30 sec and discard the supernatant.
● Repeat the washing in Step (4) once.
● Place the reaction tube containing magnetic beads on the magnetic stand, open the lid and keep it at room temperature for 5-10 min until it is dried.
Note: Do not over-dry magnetic beads, as this will cause a decrease in the yield.
● Remove the reaction tube from the magnetic stand, and thoroughly resuspend the magnetic beads by adding 22.5 μl 10 mM Tris-HCl (pH8.0) to the centrifuge tube and gently mix by up and down
for 10 times. Place the tube at room temperature for 5 min, and keep the reaction tube on the magnetic stand for 5 min. When the magnetic beads are completely attached, transfer about 20 μl of the supernatant to a new centrifuge tube for subsequent PCR amplification.
Note: Alternatively, if library amplification is not intended for the ligation product, add 12.5 μl of 10 mM Tris-HCl (pH8.0) to Step (6) for the elution of DNA, and 10 μl of the purified DNA can be transferred for subsequent application. If not proceeding immediately, please keep samples stored at -20°C.
● If size selection is needed, 102.5 μl Nuclease-Free H2O was added for elution. Then transfer about 100 μl supernatant to a new centrifuge tube for subsequent DNA size selection.
● For size selection: TIANSeq Size Selection DNA Beads (Cat# 4992358/4992359/4992979 ) is recommended. Please refer to the proportion ofmagnetic beads in the size selection process in Table 1 for operation.
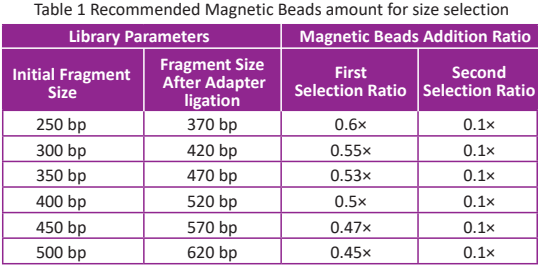
Taking the case with the initial fragment size of 250 bp as an example, when TIANSeq Size Selection DNA Beads is used for purification, the specific steps are as follows:
● Equilibrate the magnetic beads at room temperature for 20 min.
● Vortex the magnetic beads to full suspension. Add 0.6× (60μl) magnetic beads (according to the first selection ratio in Table 1) to 100 μl ligation product from Step 4 (8). Mix thoroughly by pipetting
up and down for 10 times.
● Incubate the mix for 5 min at room temperature. Place the reaction tube on the magnetic stand for 5 min. When the magnetic beads are completely attached, carefully transfer the supernatant to a new centrifuge tube containing 0.1× (10 μl) magnetic beads with a pipette (do not aspirate the beads when pipetting), and immediately pipette up and down for at least 10 times to mix thoroughly.
● Incubate the mix for 5 min at room temperature. Place the reaction tube on the magnetic stand for 5 min. When the magnetic beads are completely attached, carefully remove the supernatant with a pipette.
● Place the tube on the magnetic stand and add 200-500 μl freshly prepared 80% ethanol (the ethanol should be just enough to immerse all the beads) to the reaction tube, then gently pipette up and down for 3-5 times to wash the magnetic beads (do not disturb the beads). Pellet the magnetic beads with a magnetic stand for 30 sec and discard the supernatant.
● Repeat the washing step once.
● Place the reaction tube containing the magnetic beads on the magnetic stand, open the lid and keep it at room temperature for 5-10 min until it is dried.
Note: Do not over-dry magnetic beads, as this will cause a decrease in the yield.
● Remove the reaction tube from the magnetic stand, and thoroughly resuspend the magnetic beads by adding 22.5 μl 10 mM Tris-HCl (pH8.0) to the centrifuge tube and gently mix by pipetting up and down for 10 times. Place the tube at room temperature for 5 min, and keep the reaction tube on the magnetic stand for 5 min. When the magnetic beads are completely attached, transfer about
20 μl supernatant to a new centrifuge tube for subsequent PCR amplification.
● Library Amplification
● Thaw the 2× HiFi PCR MasterMix and P5/P7 Primers Mix (10 μM each) on ice. Once thawed, mix the 2× HiFi PCR MasterMix by finger flicking and inverting up and down. The P5/P7 Primer Mix (10 μM) can be mixed by quick vortexing.
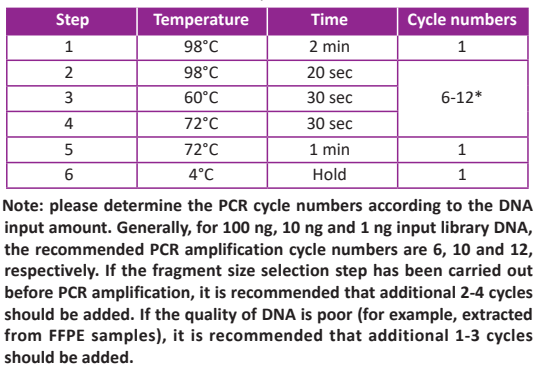
● Program a thermal cycler with the parameters listed in the table below. Turn on the hot lid and set the temperature to 105°C.
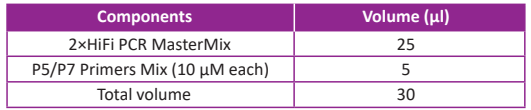
● Prepare the PCR reaction according to the following table. Note that this step needs to be operated on ice.
● Transfer 20 μl of the purified ligation product to the PCR tube. Add 30 μl PCR reaction solution prepared in Step 3 into the tube and gently mix by pipetting for 10 times.
Note: Please keep the reaction tube on ice the whole time during reaction set up.
● Pulse-spin the PCR reaction tube, and immediately transfer the tube to the thermal cycler. Start the program that's been set in Step 2.
● When the thermal cycler program is complete and sample block has returned to 4°C, remove the sample from the block and proceed to the purification step using 1× (50 μl) TIANSeq Size Selection DNA Beads (Cat# 4992358/4992359/4992979).
● Equilibrate the magnetic beads at room temperature for 20 min.
● Vortex the magnetic beads to full suspension. Add 50 μl magnetic beads to the solution. Mix thoroughly by pipetting up and down for 10 times.
● Incubate the mix for 5 min at room temperature. Place the reaction tube on the magnetic stand for 5 min. When the magnetic beads are completely attached, carefully remove the supernatant with a pipette.
● Place the tube on the magnetic stand and add 200-500 μl freshly prepared 80% ethanol (the ethanol should be just enough to immerse all the beads) to the reaction tube, then gently pipette up and down for 3-5 times to wash the magnetic beads (do not disturb the beads). Pellet the magnetic beads with a magnetic stand for 30 sec and discard the supernatant.
● Repeat the washing in Step (4) once.
● Place the reaction tube containing the magnetic beads on the magnetic stand. Open the lid to dry for 5-10 min at room temperature until it is dried.
Note: Do not over-dry magnetic beads, as this will cause a decrease in the yield.
● Remove the reaction tube from the magnetic stand, and thoroughly resuspend the magnetic beads by adding 22.5 μl of 10 mM Tris-HCl (pH8.0) to the centrifuge tube and mix the magnetic beads
thoroughly by pipetting up and down for 10 times. Place the tube at room temperature for 5 min, and keep the reaction tube on the magnetic stand for 5 min. When the magnetic beads are completely
attached to the tube wall, transfer 20 μl of the supernatant to a new centrifuge tube.
● The quality of the DNA library can be evaluated by gel electrophoresis, qPCR quantification or Agilent bioanalyzer before sending for sequencing. The purified DNA library can be stored at -20°C.
Experimental Example
-
Sort by
-
Date
Date(
)
Date
Date(
)
Impact Factor
IF(
)
Impact Factor
IF(
)



 Inquire
Inquire